How to convert data to BIDS format
The BIDS Importer. The BIDS Importer is a step-by-step tool that can be used to import raw data into a BIDS dataset.
About this tutorial
The purpose of this guide is to familiarize HIP users with the BIDS tools accessible on the platform. These tools are tailored for the platform and enable users to easily generate, populate, and oversee BIDS datasets through a step-by-step approach.
Having a fundamental understanding of the BIDS standard is crucial for utilizing the BIDS tools on the platform. Therefore, this guide initially provides an overview of the BIDS standard and its key principles. Subsequently, it delves into the BIDS tools, outlining their primary functionalities and limitations via a video guide. This guide, combined with the tips available in the BIDS tools, should should be sufficient to perform basic operation on BIDS datasets without requiring prior knowledge of the BIDS standard.
The importation process is fully guided but has a limited scalability and is time consuming. Therefore, advanced users should rely on scripting using the BIDS Manager, available on the platform, so they can programmatically prepare and import data into BIDS datasets.
BIDS at a glance
The Brain Imaging Data Structure (BIDS) is a standard for organizing, annotating, and describing neuroimaging, neurophysiological and behavioral data. It is a framework for organizing data that standardizes file organization and dataset description.
For more details regarding the standard, please refer to the following external resources:
BIDS is a community-driven and its specifications can be extended to new data types using the guide for BIDS extensions proposals . Several papers have been published covering the standard and its extensions:
BIDS basics
BIDS standardize the way to organize, name and describe files of a dataset.
In practical terms, a BIDS dataset comprises a directory that houses all the data, known as the raw directory, with a dedicated folder for each subject. Within these subject folders, there are subfolders for different data types, organizing similar data together. Data files must adhere to a specific naming convention based on BIDS entities, and only certain open-source formats are permitted. To provide additional information about the dataset and its files, metadata files are used for description.
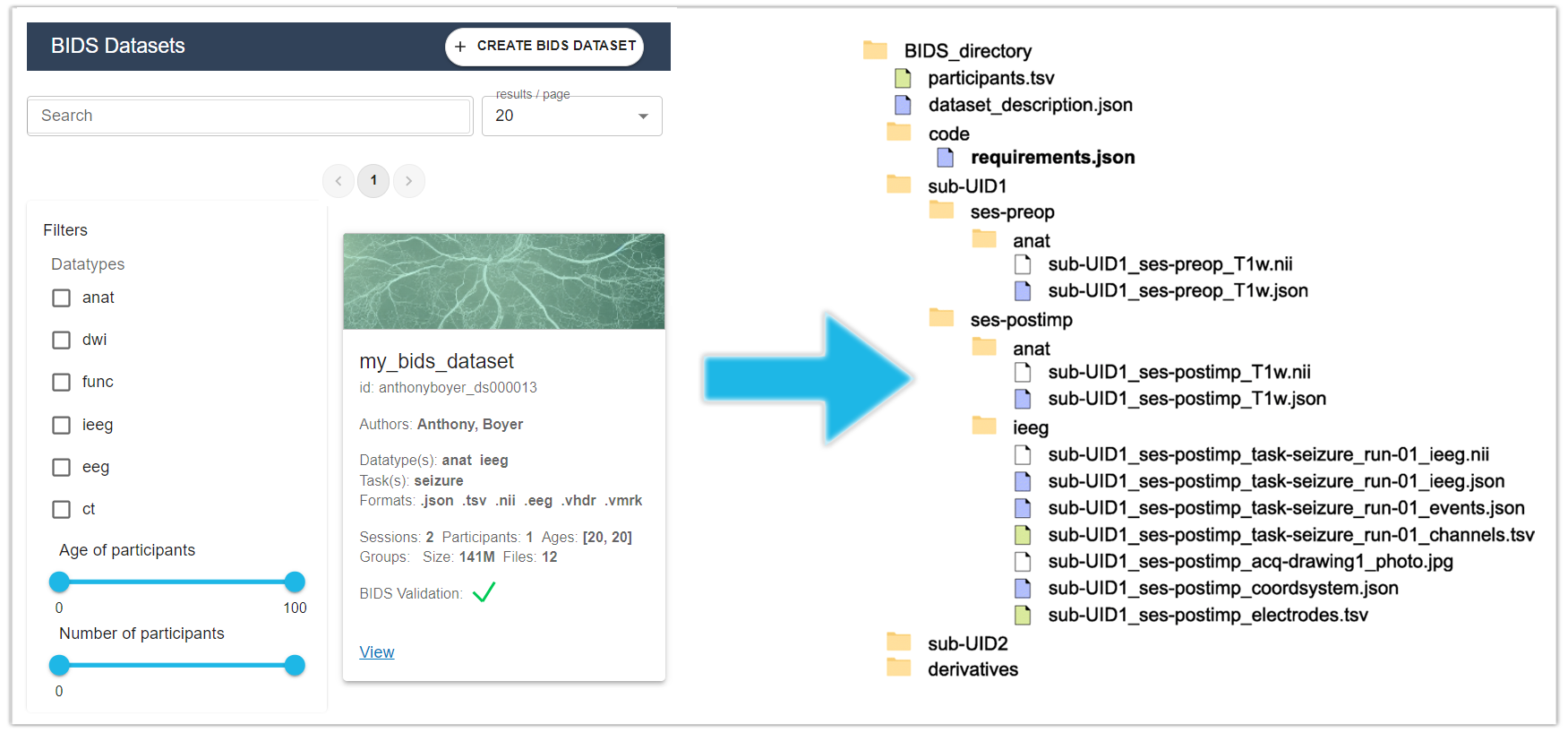
The BIDS directory structure can be summarized as follows:
The dataset name should be highly descriptive, and it’s essential for subject folders to have distinct names. Session subfolders can be employed to organize data logically, such as creating separate session subfolders for each visit by a subject in a longitudinal study. If session subfolders are utilized, they must be consistently used for all subjects to maintain uniformity in the directory structure. Data type folders are designed to group together similar acquisition modalities. However, it’s important to note that not all data types are supported by the standard, although it can be extended through BIDS extensions proposals to include additional data types.
Supported BIDS data types are as follows:
BIDS data type |
Description |
|---|---|
func |
Functional MRI data. |
dwi |
Diffusion weighted imaging data. |
fmap |
Field inhomogeneity mapping data. |
anat |
Structural imaging data. |
meg |
Magnetoencephalography data. |
eeg |
Electroencephalography data. |
ieeg |
Intracranial electroencephalography data. |
beh |
Behavioral data. |
pet |
Positron emission tomography data. |
perf |
Perfusion imaging data. |
Data files must conform to a selection of open-source formats given by the standard. Metadata files are used to describe data files or the dataset itself and are stored in companion *.json or *.tsv files.
Supported BIDS file types are as follows:
.json: contain “key: value” metadata.
.tsv: contain tables of metadata.
raw: data files in BIDS compliant format (e.g. NIfTI, BrainVision, European data format).
An important aspect of the standard is that data file names have to use BIDS entities so they are unique and informative. BIDS entities are key-value pairs chained with underscores and are used to structure file names.
BIDS file naming convention is illustrated in the figure below:
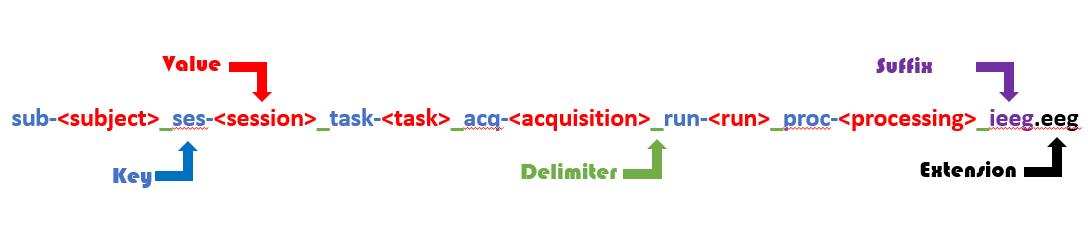
BIDS file naming. File names are structured using BIDS entities (key-value pairs) connected with underscores and ending with a suffix and a file extension.
BIDS entities are used at the subject level on raw data and associated metadata files, between- and within- subjects, but may not be used on some study-level metadata files (e.g. dataset_description.json, participants.tsv, *sessions.tsv, *scans.tsv)
BIDS entities have a definite order. Keys are alphanumeric while values can be either alphanumeric or integers depending on the considered entity. BIDS entities can be required or optional, depending on the modality and design choices of the BIDS dataset, and some have to be consistent across subjects/sessions when used.
The following table is an example of BIDS entities which can be found in a BIDS dataset:
Entity |
Key |
Description |
Example |
|---|---|---|---|
Subject |
sub |
A person or animal participating in the study. |
sub-johndoe |
Session |
ses |
An intermediate folder in BIDS folder hierarchy to group data that go “logically” together. If used, sessions must be pertinent and consistent across subjects. |
ses-postsurgery |
Task |
task |
Identify the task performed by the subject during the acquisition. If used, must be consistent across subjects and sessions. |
task-eyesclosed |
Acquisition |
acq |
Identify the acquisition parameters used to perform the acquisition. If used, must be consistent across subjects and sessions. |
acq-lowres |
Contrast enhancing agent |
ce |
Identify contrast agent. Does not have to be consistent. |
ce-gadolinium |
Reconstruction |
rec |
Identify reconstruction algorithms. Does not have to be consistent. |
rec-lsqr |
Phase-Encoding Direction |
dir |
Identify phase-encoding directions. Does not have to be consistent. |
dir-leftright |
Run |
run |
Identify repeated acquisitions. Does not have to be consistent. But becomes required if more than one acquisition. Value is an integer. |
run-01 |
Modality |
mod |
Identify modality label for defacing masks. Does not have to be consistent. |
mod-T1w_defacemask |
Echo |
echo |
Identify an echo file with its index. Does not have to be consistent. Value is an integer. |
echo-01 |
Recording |
recording |
Identify different continuous recording files. Does not have to be consistent. |
recording-saturation |
Processed |
proc |
Identify processing applied during acquisition. Does not have to be consistent. |
proc-trans |
Space |
space |
Identify the space in which coordinates/positions should be interpreted. Does not have to be consistent. Value come from dedicated BIDS appendix VIII: Coordinate systems. |
space-MNI152Lin |
Split |
split |
Identify multiple parts of a file. Mostly used for .fif files. Does not have to be consistent. Value is an integer. |
split-01 |
Tracer |
trc |
Identify the radio tracer used during PET acquisition. |
trc-fdg |
Unsupported data files
Data files that are not covered by the standard may still be imported in a BIDS dataset. They should follow BIDS guidelines as much as possible and have to be declared in a .bidsignore file in BIDS raw directory in order to pass BIDS validation. This is not supported by the BIDS tools available on the platform.
Derivatives files
Data files which have been processed and that cannot be considered as raw data anymore should go to the BIDS derivatives folder. This is not supported by the BIDS tools available on the platform.
BIDS tools
The BIDS tools are custom-built to facilitate the straightforward creation, population, and management of BIDS datasets, employing a user-friendly, step-by-step process. These tools prioritize simplicity, readability, and data integrity. Their actions are intentionally limited to prevent unintended changes. For instance, the importation tool is constrained to essential BIDS entities, and most metadata files cannot be directly edited through the interface, with only a few exceptions.
Supported data types and corresponding BIDS modalities are as follows:
BIDS data type |
BIDS modalities |
|---|---|
anat |
T1w, T2w, T1rho, T1map, T2map, T2start, FLAIR, PD, Pdmap, PDT2, inplanteT1, inplanteT2, angio, defacemask, CT |
pet |
pet, petmr, petct |
func |
bold, sbref |
fmap |
phasediff, phase1, phase2, magnitude, magnitude1, magnitude2, fieldmap, epi |
dwi |
dwi |
ieeg |
ieeg |
eeg |
eeg |
meg |
meg |
ieegGlobalSidecars |
electrodes, coordsystem, photo |
eegGlobalSidecars |
electrodes, coordsystem, photo |
petGlobalSidecars |
blood |
Supported data file formats are as follows:
File format |
Extension |
|---|---|
Micromed |
.trc (.trc) |
BrainVision |
.vhdr, .vmrk, .eeg (.vhdr) |
European data format |
.edf (.edf) |
BioSemi data format |
.bdf (.bdf) |
EEGLAB files |
.set (.set) |
DICOM |
.dcm (root directory of .dcm files) |
NifTi |
.nii (.nii) |
Video guide
The following video guide (6’09’’) serves as an introduction to the BIDS tools and shows how to import raw data into a new BIDS dataset:
References
Gorgolewski, K., Auer, T., Calhoun, V. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3, 160044 (2016).
Gorgolewski KJ, Alfaro-Almagro F, Auer T, Bellec P, Capotă M, Chakravarty MM, et al. BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput Biol 13(3) (2017).
Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R. EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci Data. 2019 Jun 25;6(1):103.
Niso, G., Gorgolewski, K., Bock, E. et al. MEG-BIDS, The brain imaging data structure extended to magnetoencephalography. Sci Data 5, 180110 (2018).
Holdgraf, Chris et al. “BIDS-iEEG: an extension to the brain imaging data structure (BIDS) specification for human intracranial electrophysiology.” (2018).
Clara A Moreau, Martineau Jean-Louis, Ross Blair, Christopher J Markiewicz, Jessica A Turner, Vince D Calhoun, Thomas E Nichols, Cyril R Pernet, The genetics-BIDS extension: Easing the search for genetic data associated with human brain imaging, GigaScience, Volume 9, Issue 10 (2020).