HOW TO
Create a HIP account
The HIP is fully integrated in EBRAINS and an EBRAINS account is required in order to access the HIP and use all of its services.
Terms of Use
By applying for a HIP account, future HIP Users accept the EBRAINS General Terms of Use, so to indicate acceptance and compliance with all applicable laws, regulations, rules and approvals in the use and sharing of the data on EBRAINS, including, but not limited to, the General Data Protection Regulation (GDPR). They also adhere to the HIP Charter which defines the principles, rules and processes that govern the management of the data and research outputs.
Create an EBRAINS account
EBRAINS is a sustainable European Research Infrastructure providing tools and services in brain research and brain-inspired technologies created by the EU-funded Human Brain Project. Part of EBRAINS resources are available publicly without an account. Other resources, such as the HIP, are accessible only to registered users.
Before applying for a HIP account you need to register for an EBRAINS account. New users with a personal institutional e-mail from a pre-identified institution will be provided an expedited registration process. If the institution is not yet on EBRAINS list, you can either contact EBRAINS High Level Support Team (HLST) or fill in the dedicated form. If you cannot provide an institutional e-mail, it is still possible to request an EBRAINS account from a personal e-mail with a short motivation (max 100 words).
Create a HIP account
You need an EBRAINS account to apply for a HIP account.
All HIP users are member of at least one group corresponding to pre-identified institutions (laboratory, hospital, university…) listed in the following table:
HIP group |
Provider |
Group leader |
|---|---|---|
AMU-NS |
Aix-Marseille University, Institut de Neurosciences des Systèmes, Neurostimulation team, France |
Dr Olivier David |
AMU-TNG |
Aix-Marseille University, Institut de Neurosciences des Systèmes, Theoretical Neurosciences Group, France |
Dr Viktor Jirsa |
APHM |
University Hospitals of Marseille, Epilepsy department, France |
Prof. Fabrice Bartolomei |
CHRU-LILLE |
Centre Hospitalier Universitaire de Lille, Epilepsy unit, France |
Prof. Philippe Derambure |
CHU-LYON |
Hospices Civils de Lyon - Centre Hospitalier Universitaire, France |
Prof. Alexis Arzimanoglou |
CHUV |
Centre Hospitalier Universitaire de Lausanne, Switzerland |
Prof. Philippe Ryvlin |
HUS |
Helsinki University Hospital, Hospital Disctrict of Helsinki and Uusimaa, Finland |
Prof. Eeva-Liisa Metsähonkala |
OU-SSE |
The Norwegian National Unit for Epilepsy, Oslo universitetssykehus, Norway |
Prof. Morten Lossius |
PSMAR |
Hospital del Mar-Parc de Salut Mar, Spain |
Prof. Rodrigo Rocamora Zuniga |
SAUH |
St. Anne’s University Hospital, Czech Republic |
Prof. Milan Brazdil |
UCBL |
Université Claude Bernard Lyon 1, France |
Dr Jean-Philippe Lachaux |
UMCU |
University Medical Center Utrecht (Brain Center Rudolf Magnus), Netherlands |
Prof. Kees Braun |
Unlisted institutions
Access to the HIP is restricted to users from registered institutions. For more information please contact the HIP support.
To register for a HIP account please send an e-mail to HIP support with the following information (items marked with an asterisk are mandatory):
Full name*: First name and surname.
HIP group(s)*: At least one HIP group you are affiliated with.
EBRAINS username*: The username you use when logging into EBRAINS services.
Institutional e-mail*: The e-mail that will be used by the HIP support team to contact you.
Backup e-mail*: A backup e-mail.
ORCID: Open Researcher and Contributor ID.
Once your application has been validated, a confirmation e-mail will be sent to your institutional e-mail and you will be added to the HIP group on EBRAINS. You can then connect to the HIP portal and access its services, logging in with your EBRAINS account, and have access to the shared folder of each group you belong to.
Accounts and groups
HIP users are not authorized to create a second account (1 EBRAINS account = 1 HIP account) but can be part of multiple groups. HIP users may contact the HIP support team to request group changes (please use the institutional e-mail you specified during the creation of your HIP account and indicate your EBRAINS username).
How to connect to the HIP portal and access its services
Connect to the HIP
The HIP is a w`b-based platform which can be accessed using any recent web browser. For the best experience with the HIP web interface, we recommend that you use the latest and supported version of a browser from the following list:
Microsoft Edge
Mozilla Firefox
Google Chrome/Chromium
Apple Safari
User experience
The HIP is an online platform designed for desktop uses, a stable and high-performance internet connection is strongly recommended for the best experience.
Once your HIP account has been created, access the HIP web portal and log in with EBRAINS (use your EBRAINS credentials), you will be automatically redirected to the HIP home page.
HIP services
Home page

Home page. The HIP home page provides a set of services (1: Files, 2: Talk, 3: Forms, 4: HIP) which come with their own interface (menu and view) that can be searched (5). The HIP account can be managed using the profile (8), contacts (7) and notifications (6) buttons.
When connecting to the HIP you are automatically redirected to the home page from which you can access the platform main features using the side navigation menu:
Dashboard: Displays various information regarding your institution and its members to whom you can send messages using Talk. The Dashboard also shows an overview of the opened Desktops and available BIDS databases.
Desktops: Desktops operate as remote virtual computers where HIP users can run applications from the App Catalog to process data uploaded or shared in their Private Space.
BIDS: The BIDS importer is a step-by-step tool that can be used to import raw data into a BIDS database.
About: The HIP home page. It contains links to the HIP website, the onboarding guide and the feedback forms.
App Catalog: A comprehensive list of all the applications made available to the HIP users so they can process their data.
Documentation: The documentation page contains links to the user documentation, the technical documentation and the HIP website.
Bug Report: A direct link to the bug report form.
Feedback: A direct link to the feedback form.
Centers: This section lists all the institutions currently participating in the HIP as well as their respective members to whom you can send messages using Talk.
The HIP offers several services, all available at the top of the home page:
Files: Browse the folders and files available in your Private Space.
Talk: Discuss with other HIP members (HIP users and the support team).
Forms: Create a new form to share with others or submit your feedback to the HIP team.
HIP: The HIP home page.
Forms
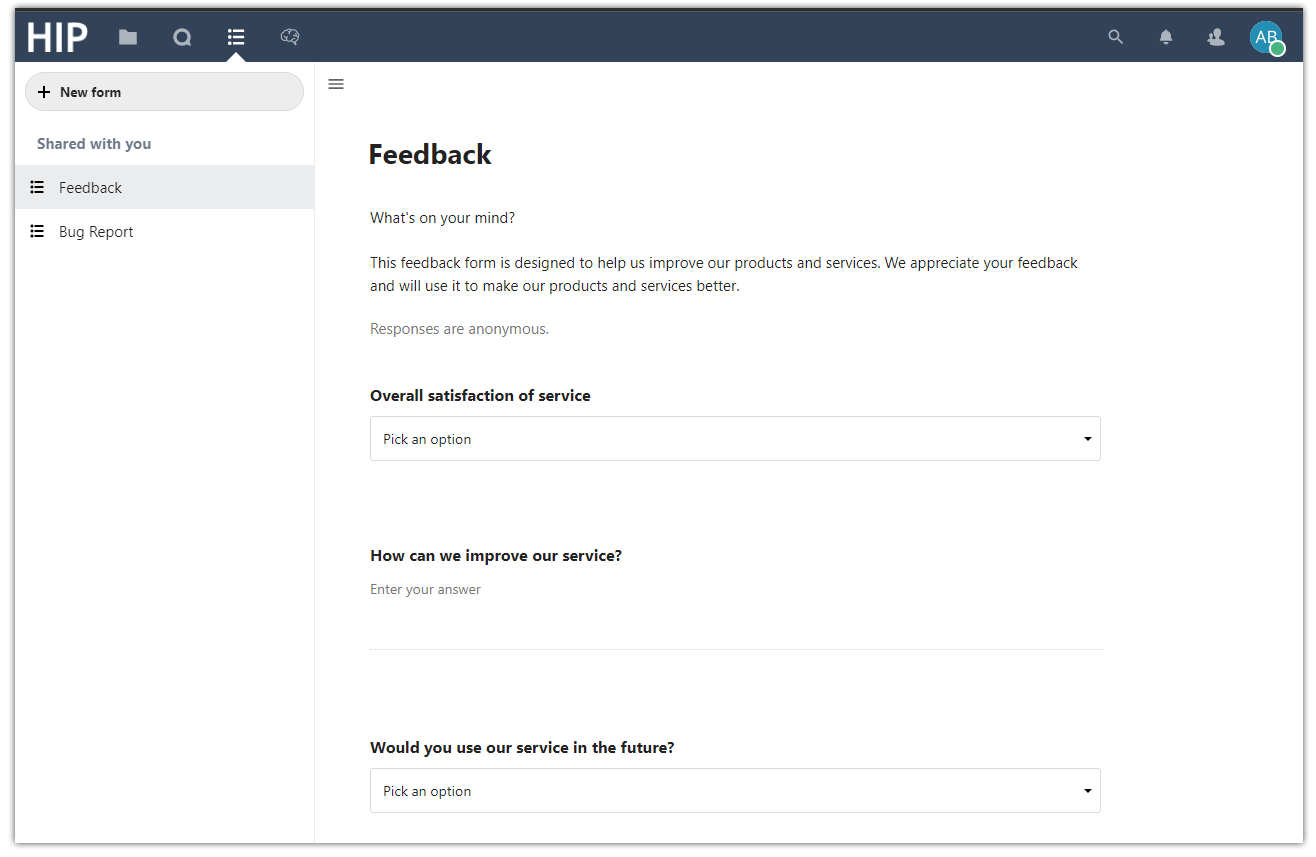
Forms. Create custom forms to share with others and give us feedback on the platform.
The Forms service lets you create custom forms (e.g. using checkboxes, dropdown lists, short answers) that you can restrict to some HIP users (specific users or groups) or share publicly (via a share link) and consult the results.
2 forms are currently available for all HIP users:
Feedback form: Designed to help us improve our products and services. We appreciate your feedback and will use it to make our products and services better.
Bug report form: Designed to help us fix bugs on the platform. Please include as much detail as possible, including steps to reproduce the bug, and what you expected to happen.
Talk
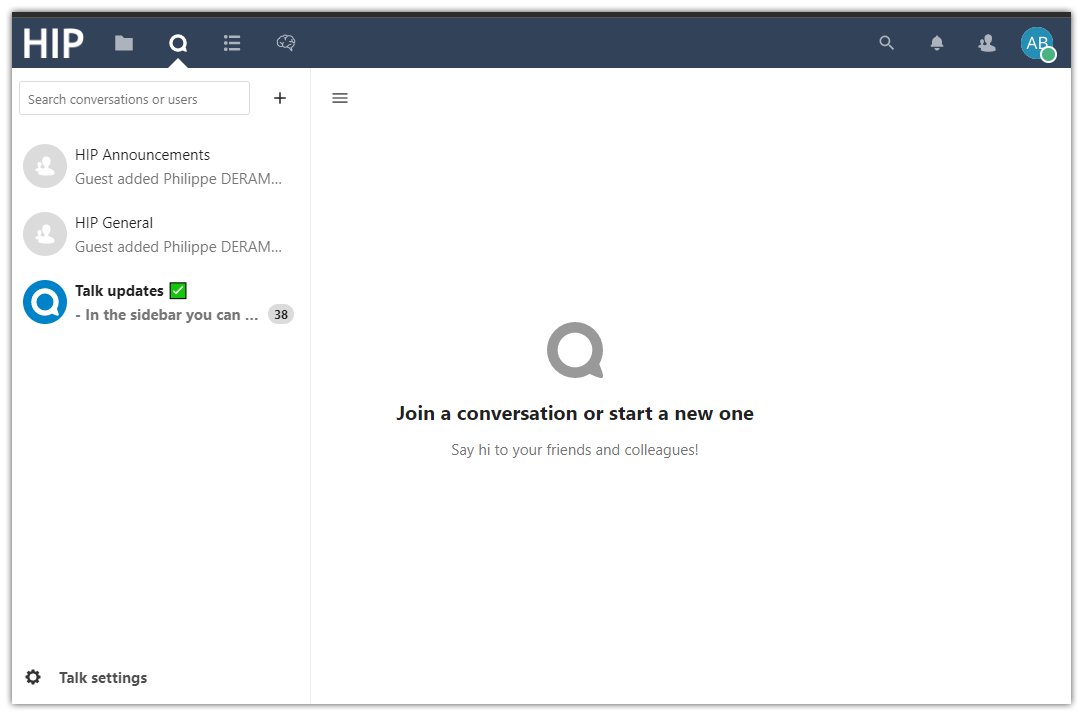
Talk. Discuss with other HIP users, group members and the HIP support team.
Use the Talk service to create your own conversation, choose its visibility, and invite other HIP users (individual users or groups of users) or guests using a link that you can protect with a password. At any time, you can select a conversation and see its members, the attachments that have been shared and saved in your Private Space, inside the “/Talk” folder. Click the “Talk settings” at the bottom left of the page to configure your webcam and microphone so you can “Start a call” with the members of the active conversation. Similarly, click on the “…” button next to any conversation to access its settings and manage notifications.
There are currently 3 conversations visible to all HIP users:
HIP Announcements: Locked conversation for important announcements regarding the HIP.
HIP General: Global conversation open to all HIP users.
Talk updates: News and patch notes regarding the Talk service.
Files
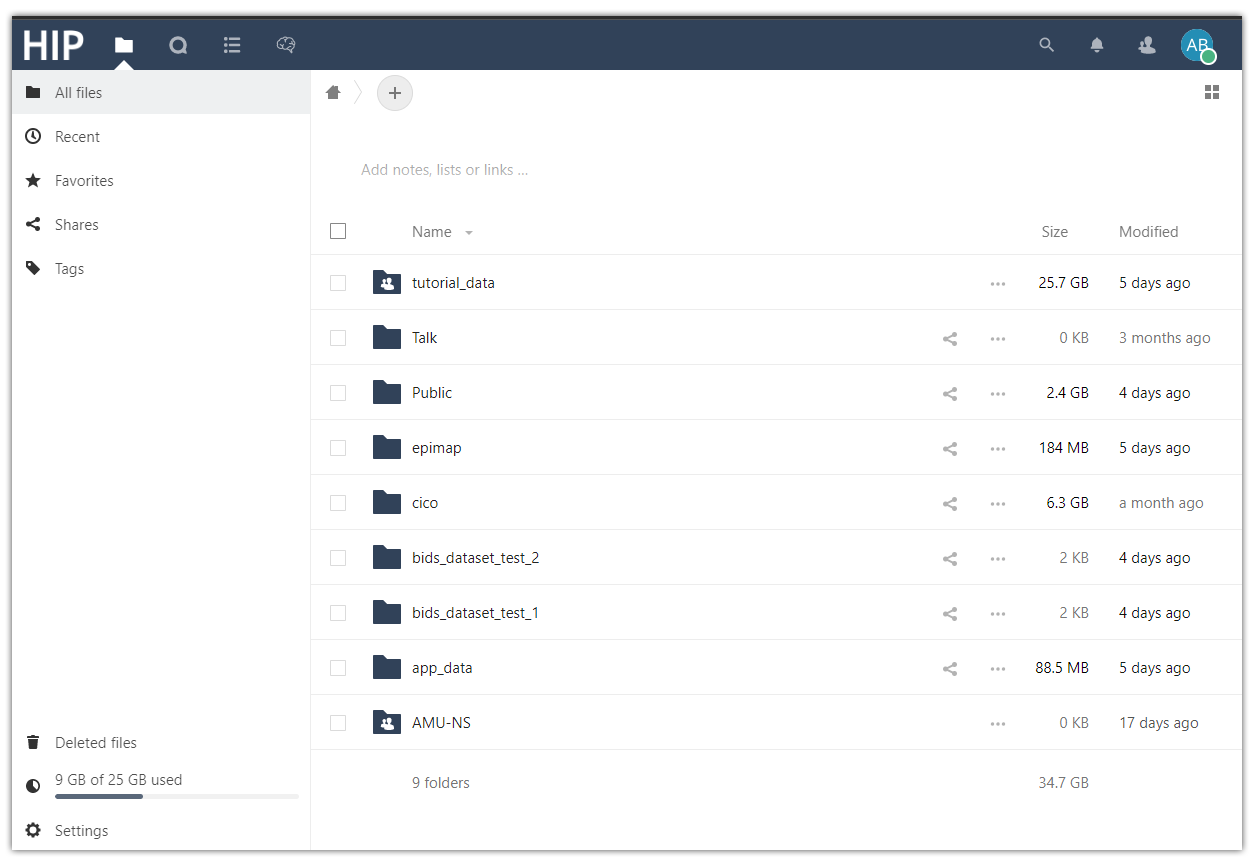
Files. Browse the folders and files available in your Private Space.
The Files service lets you browse the folders and files available in your Private Space. You can upload data to your Private Space by simply dragging and dropping files into the file browser, although it is advised to upload data using the Nexclout client as explained in the How to prepare and upload data to the HIP guide. The Private Space also contains the shared folders of the HIP groups you are a member of.
Video guide
The following video (2’15’’) shows the connection procedure and gives an overview of HIP services.
How to prepare and upload data to the HIP
Upload data to the HIP. Upload data directly from the web browser or using the Nextcloud client.
A Data Transfer Agreement (DTA) has to be signed between the participating institution and the CHUV prior to any data transfer taking place.
Data can only be uploaded in the Private Space of the HIP user doing the transfer. The Private Space enables the HIP user to curate the data to make them eligible for collaboration. Transferring data to the Collaborative or Public Spaces requires to adhere to the specifics of those spaces which are detailed in the how to use the HIP spaces and share data with other users guide. It is not possible to upload or download data directly to or from the Collaborative or Public Spaces.
Data stored on the HIP
The HIP iEEG data is organized according to the FAIR principles within the ethics and regulatory frameworks that apply to medical personal data. User and application data on the HIP are primarily stored as files and folders either in the Private, Collaborative and Public Spaces. Please refer to the associated guide for more information regarding the HIP spaces.
Before uploading data to the HIP
The HIP is an open-source European platform dedicated to Human intracerebral EEG data and only iEEG data and relevant health-related or research-related data should be uploaded to the platform.
This includes but is not limited to:
iEEG raw data, as well as the corresponding neuroimaging raw data used to identify the anatomical location of iEEG electrodes, all collected during clinical activities.
Health-related data collected during clinical activities such as patients’ characteristics (e.g. age, gender, MRI findings, localization of the epileptogenic zone) and anatomical localization of iEEG recording leads.
Research-related data specifically collected for a research purpose during iEEG (e.g. the COGEPISTIM dataset), and typically informing on the behavioral performance of patients during the research protocol (e.g. seizure recording, functional electrical stimulation, neuroanatomical imaging, cognitive research tasks or response time).
Data types and formats are not monitored or controlled, primarily due to the security principles of the HIP. Do not upload non-iEEG related data as it goes against HIP policy and such actions might be referred.
International HIP Registry
All HIP data are included in the international HIP Registry coordinated by the CHUV Project Leader and Management Team, providing a sustained regulatory framework for collecting and storing these data.
The Data Controller responsibilities
The HIP user uploading data fr`m patients onto the HIP is affiliated with at least one of the HIP groups and will qualify as Data Controller for the corresponding data.
It is the Data Controller responsibility to secure proper data anonymization. Data has to be pseudonymised/anonymised prior to being stored on the HIP platform depending on the DTA.
Data will be uploaded in the Data Controller’s Private Space, where they cannot be shared with HIP users from other iEEG centers, yet can be processed with all available HIP functionalities.
File formats
The HIP primarily relies on BIDS as a common format and database and it is strongly recommended to upload data following BIDS guidelines. For more information regarding the BIDS standard, please refer to the Brain Imaging Data Structure section of the How to convert data to BIDS format guide.
Uploaded data can be processed with all available HIP functionalities but not be shared in the Collaborative of Public Space unless they adhere to the specifics of those spaces which include BIDS validation.
The HIP provides a fully integrated tool called BIDS importer which can be used to import the uploaded data into a (new) BIDS database, taking care of data pseudonymization, normalization of formats, samplings, coordinates and other relevant metadata. Note that the BIDS importer can only convert a limited number of input formats and covers a subset of BIDS data types.
Uploading data to the HIP
There are currently 2 solutions for uploading data to the HIP: either directly from the web browser or using the Nextcloud client. It is advised to use the Nextcloud client as it uses WebDAV protocol, a more robust way to upload large/numerous files.
Using the web browser
It is possible to upload data by simply selecting the files and folders to transfer and drag and drop them into the web browser while the Private Space web page is opened. Alternatively, it is possible to use the dedicated Upload file button on the said web page to upload data. Both solutions are illustrated in the video clips below:
Note that closing the web browser before the uploading/downloading process is over will abort the operation and the transfer will not resume where it stopped.
Using the Nextcloud client
The Nextcloud client uses the WebDAV protocol to seamlessly synchronize data between the HIP user’s Private Space and the local desktop where the client is installed.
The following video guide (2’27’’) shows how to download, install and configure the Nextcloud client to set up a synchronized folder:
How to convert data to BIDS format
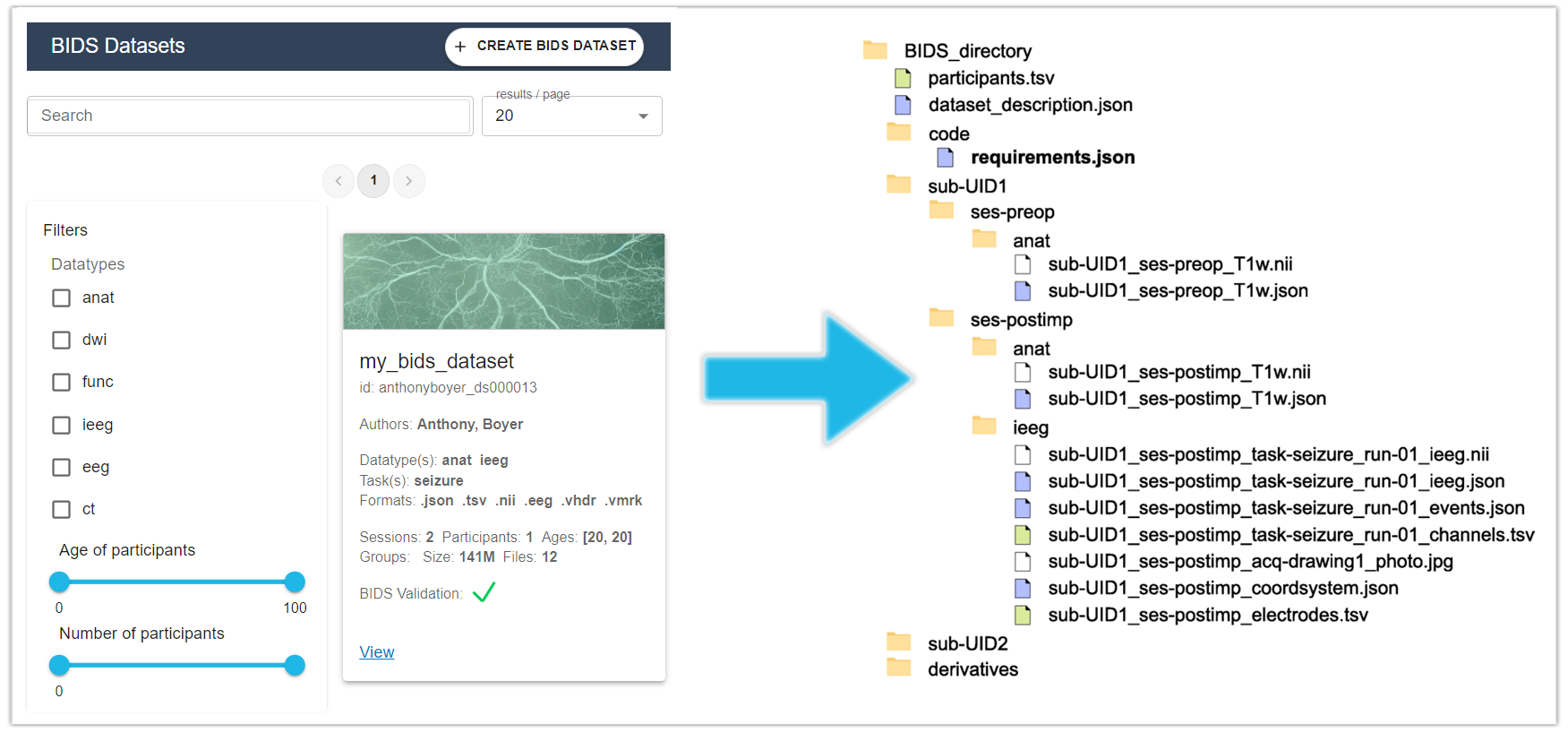
The BIDS Importer. The BIDS Importer is a step-by-step tool that can be used to import raw data into a BIDS dataset.
About this tutorial
The purpose of this guide is to familiarize HIP users with the BIDS tools accessible on the platform. These tools are tailored for the platform and enable users to easily generate, populate, and oversee BIDS datasets through a step-by-step approach.
Having a fundamental understanding of the BIDS standard is crucial for utilizing the BIDS tools on the platform. Therefore, this guide initially provides an overview of the BIDS standard and its key principles. Subsequently, it delves into the BIDS tools, outlining their primary functionalities and limitations via a video guide. This guide, combined with the tips available in the BIDS tools, should should be sufficient to perform basic operation on BIDS datasets without requiring prior knowledge of the BIDS standard.
The importation process is fully guided but has a limited scalability and is time consuming. Therefore, advanced users should rely on scripting using the BIDS Manager, available on the platform, so they can programmatically prepare and import data into BIDS datasets.
BIDS at a glance
The Brain Imaging Data Structure (BIDS) is a standard for organizing, annotating, and describing neuroimaging, neurophysiological and behavioral data. It is a fra`ework for organizing data that standardizes file organization and dataset description.
For more details regarding the standard, please refer to the following external resources:
BIDS is a community-driven and its specifications can be extended to new data types using the guide for BIDS extensions proposals . Several papers have been published covering the standard and its extensions:
BIDS basics
BIDS standardize the way to organize, name and describe files of a dataset.
In practical terms, a BIDS dataset comprises a directory that houses all the data, known as the raw directory, with a dedicated folder for each subject. Within these subject folders, there are subfolders for different data types, organizing similar data together. Data files must adhere to a specific naming convention based on BIDS entities, and only certain open-source formats are permitted. To provide additional information about the dataset and its files, metadata files are used for description.
The BIDS directory structure can be summarized as follows:
The dataset name should be highly descriptive, and it’s essential for subject folders to have distinct names. Session subfolders can be employed to organize data logically, such as creating separate session subfolders for each visit by a subject in a longitudinal study. If session subfolders are utilized, they must be consistently used for all subjects to maintain uniformity in the directory structure. Data type folders are designed to group together similar acquisition modalities. However, it’s important to note that not all data types are supported by the standard, although it can be extended through BIDS extensions proposals to include additional data types.
Supported BIDS data types are as follows:
BIDS data type |
Description |
|---|---|
func |
Functional MRI data. |
dwi |
Diffusion weighted imaging data. |
fmap |
Field inhomogeneity mapping data. |
anat |
Structural imaging data. |
meg |
Magnetoencephalography data. |
eeg |
Electroencephalography data. |
ieeg |
Intracranial electroencephalography data. |
beh |
Behavioral data. |
pet |
Positron emission tomography data. |
perf |
Perfusion imaging data. |
Data files must conform to a selection of open-source formats given by the standard. Metadata files are used to describe data files or the dataset itself and are stored in companion *.json or *.tsv files.
Supported BIDS file types are as follows:
.json: contain “key: value” metadata.
.tsv: contain tables of metadata.
raw: data files in BIDS compliant format (e.g. NIfTI, BrainVision, European data format).
An important aspect of the standard is that data file names have to use BIDS entities so they are unique and informative. BIDS entities are key-value pairs chained with underscores and are used to structure file names.
BIDS file naming convention is illustrated in the figure below:
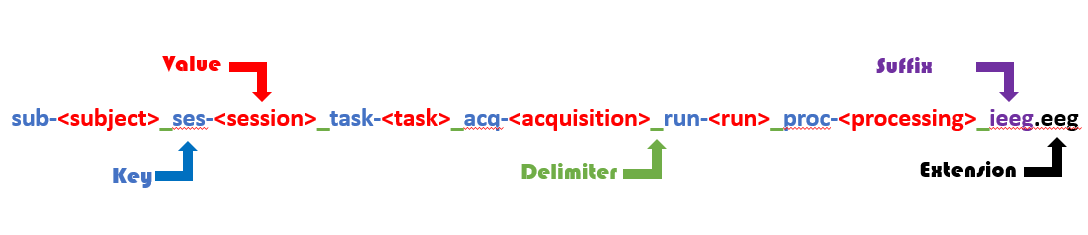
BIDS file naming. File names are structured using BIDS entities (key-value pairs) connected with underscores and ending with a suffix and a file extension.
BIDS entities are used at the subject level on raw data and associated metadata files, between- and within- subjects, but may not be used on some study-level metadata files (e.g. dataset_description.json, participants.tsv, *sessions.tsv, *scans.tsv)
BIDS entities have a definite order. Keys are alphanumeric while values can be either alphanumeric or integers depending on the considered entity. BIDS entities can be required or optional, depending on the modality and design choices of the BIDS dataset, and some have to be consistent across subjects/sessions when used.
The following table is an example of BIDS entities which can be found in a BIDS dataset:
Entity |
Key |
Description |
Example |
|---|---|---|---|
Subject |
sub |
A person or animal participating in the study. |
sub-johndoe |
Session |
ses |
An intermediate folder in BIDS folder hierarchy to group data that go “logically” together. If used, sessions must be pertinent and consistent across subjects. |
ses-postsurgery |
Task |
task |
Identify the task performed by the subject during the acquisition. If used, must be consistent across subjects and sessions. |
task-eyesclosed |
Acquisition |
acq |
Identify the acquisition parameters used to perform the acquisition. If used, must be consistent across subjects and sessions. |
acq-lowres |
Contrast enhancing agent |
ce |
Identify contrast agent. Does not have to be consistent. |
ce-gadolinium |
Reconstruction |
rec |
Identify reconstruction algorithms. Does not have to be consistent. |
rec-lsqr |
Phase-Encoding Direction |
dir |
Identify phase-encoding directions. Does not have to be consistent. |
dir-leftright |
Run |
run |
Identify repeated acquisitions. Does not have to be consistent. But becomes required if more than one acquisition. Value is an integer. |
run-01 |
Modality |
mod |
Identify modality label for defacing masks. Does not have to be consistent. |
mod-T1w_defacemask |
Echo |
echo |
Identify an echo file with its index. Does not have to be consistent. Value is an integer. |
echo-01 |
Recording |
recording |
Identify different continuous recording files. Does not have to be consistent. |
recording-saturation |
Processed |
proc |
Identify processing applied during acquisition. Does not have to be consistent. |
proc-trans |
Space |
space |
Identify the space in which coordinates/positions should be interpreted. Does not have to be consistent. Value come from dedicated BIDS appendix VIII: Coordinate systems. |
space-MNI152Lin |
Split |
split |
Identify multiple parts of a file. Mostly used for .fif files. Does not have to be consistent. Value is an integer. |
split-01 |
Tracer |
trc |
Identify the radio tracer used during PET acquisition. |
trc-fdg |
Unsupported data files
Data files that are not covered by the standard may still be imported in a BIDS dataset. They should follow BIDS guidelines as much as possible and have to be declared in a .bidsignore file in BIDS raw directory in order to pass BIDS validation. This is not supported by the BIDS tools available on the platform.
Derivatives files
Data files which have been processed and that cannot be considered as raw data anymore should go to the BIDS derivatives folder. This is not supported by the BIDS tools available on the platform.
BIDS tools
The BIDS tools are custom-built to facilitate the straightforward creation, population, and management of BIDS datasets, employing a user-friendly, step-by-step process. These tools prioritize simplicity, readability, and data integrity. Their actions are intentionally limited to prevent unintended changes. For instance, the importation tool is constrained to essential BIDS entities, and most metadata files cannot be directly edited through the interface, with only a few exceptions.
Supported data types and corresponding BIDS modalities are as follows:
BIDS data type |
BIDS modalities |
|---|---|
anat |
T1w, T2w, T1rho, T1map, T2map, T2start, FLAIR, PD, Pdmap, PDT2, inplanteT1, inplanteT2, angio, defacemask, CT |
pet |
pet, petmr, petct |
func |
bold, sbref |
fmap |
phasediff, phase1, phase2, magnitude, magnitude1, magnitude2, fieldmap, epi |
dwi |
dwi |
ieeg |
ieeg |
eeg |
eeg |
meg |
meg |
ieegGlobalSidecars |
electrodes, coordsystem, photo |
eegGlobalSidecars |
electrodes, coordsystem, photo |
petGlobalSidecars |
blood |
Supported data file formats are as follows:
File format |
Extension |
|---|---|
Micromed |
.trc (.trc) |
BrainVision |
.vhdr, .vmrk, .eeg (.vhdr) |
European data format |
.edf (.edf) |
BioSemi data format |
.bdf (.bdf) |
EEGLAB files |
.set (.set) |
DICOM |
.dcm (root directory of .dcm files) |
NifTi |
.nii (.nii) |
Video guide
The following video guide (6’09’’) serves as an introduction to the BIDS tools and shows how to import raw data into a new BIDS dataset:
Reference
Gorgolewski, K., Auer, T., Calhoun, V. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3, 160044 (2016).
Gorgolewski KJ, Alfaro-Almagro F, Auer T, Bellec P, Capotă M, Chakravarty MM, et al. BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput Biol 13(3) (2017).
Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R. EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci Data. 2019 Jun 25;6(1):103.
Niso, G., Gorgolewski, K., Bock, E. et al. MEG-BIDS, The brain imaging data structure extended to magnetoencephalography. Sci Data 5, 180110 (2018).
Holdgraf, Chris et al. “BIDS-iEEG: an extension to the brain imaging data structure (BIDS) specification for human intracranial electrophysiology.” (2018).
Clara A Moreau, Martineau Jean-Louis, Ross Blair, Christopher J Markiewicz, Jessica A Turner, Vince D Calhoun, Thomas E Nichols, Cyril R Pernet, The genetics-BIDS extension: Easing the search for genetic data associated with human brain imaging, GigaScience, Volume 9, Issue 10 (2020).
How to use Desktops and run applications from the App Catalog
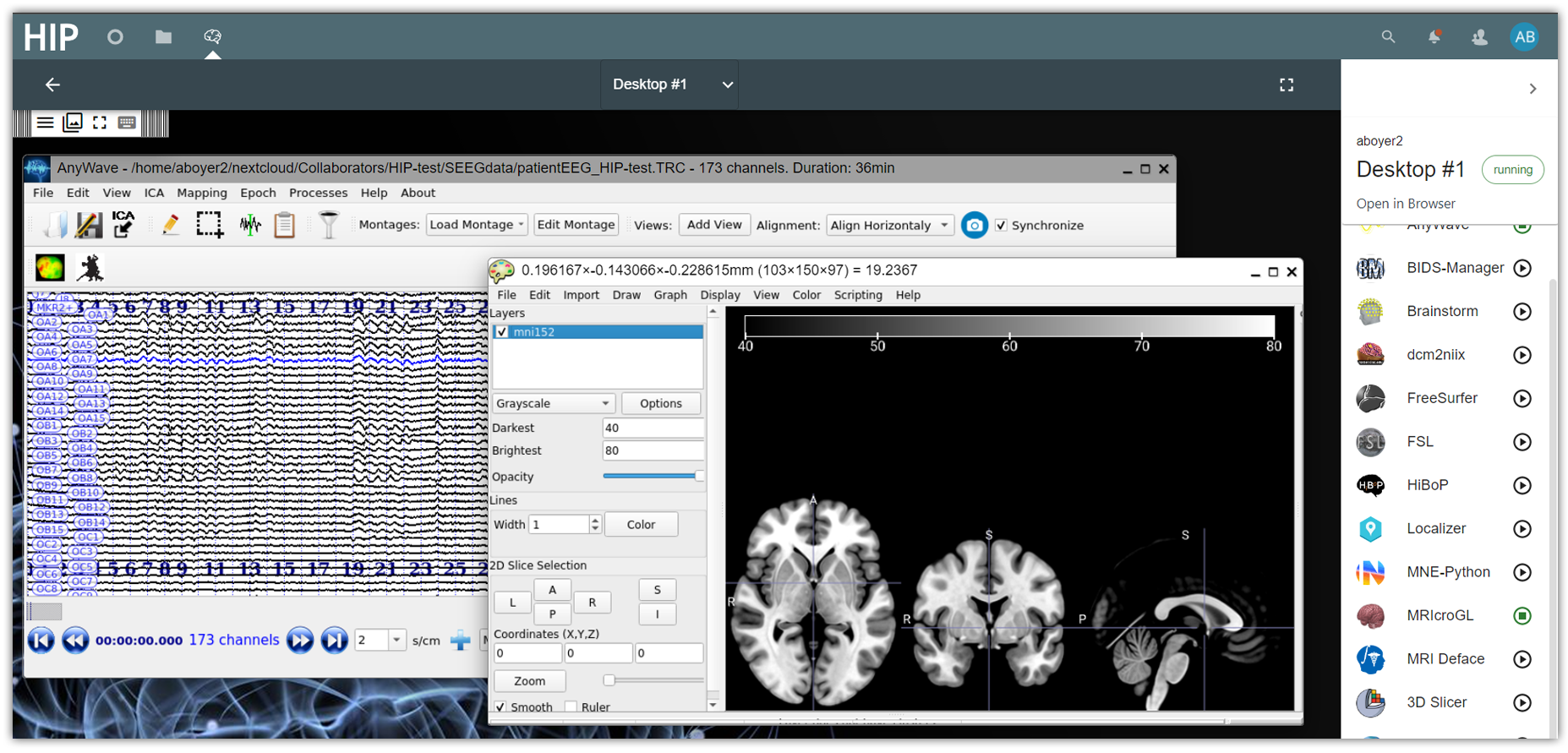
Desktops on the HIP. Desktops are persistent environments used to run applications from the App Catalog.
About this tutorial
The objective of this guide is to introduce Desktops, which operate as remote computers and are typically used to run the applications available in the App Catalog. This guide comes with a video showing how to manage Desktops (create, display, pause/resume and switch between Desktops) and run one or several applications simultaneously. It also explains essential rules regarding data access and persistence inside a Desktop environment. However, it does not cover the uses of the various applications available in the App Catalog. In this perspective, the HIP APPS and HIP TUTORIALS sections accessible in the table of contents contain documentation, tutorials and links to external resources.
Use Desktops and run applications from the App Catalog
Desktops are remote virtual computers running on a secure infrastructure where HIP users can run applications from the App Catalog to work on their data. Desktops operate similarly to a personal computer and support most common features (drag and drop, full screen mode, clipboard, virtual keyboard). Any application from the App Catalog, regardless if it is GUI- or CLI- based, can be run in a Desktop environment. Desktops are available in all 3 HIP spaces: the Private, Collaborative and Public spaces, and work in a similar way.
Desktops and data persistence
Once it has been initiated, a Desktop will persist until it is manually terminated. HIP users can safely log off and/or close their web browser. Pending Desktops will remain unaltered and accessible for later use.
Important
Applications running in a Desktop environment have access to your Private Space data under the /home/<HIP_USER>/nextcloud directory. Any data and/or configuration file outside of this directory will be lost when the application or desktop are closed. This is the only persistent directory as it is tied to the HIP user’s Private Space at application startup. Make sure you are always working within the /home/<HIP_USER>/nextcloud directory when using applications.
Video guide
The following video guide (2’30’’) serves as an introduction to Desktops and shows how to run applications from the App Catalog:
How to transfer data from Micromed software to the HIP
It is possible to seamlessly synchronize a database created with Micromed software (SystemPLUS EVOLUTION software) so it is available in the HIP user’s Private Space. This requires to set up a synchronized folder using the Nextcloud client as explained in the How to prepare and upload data to the HIP guide and then create the database in said folder.
Once the Nextcloud synchronized folder is ready, take the following steps:
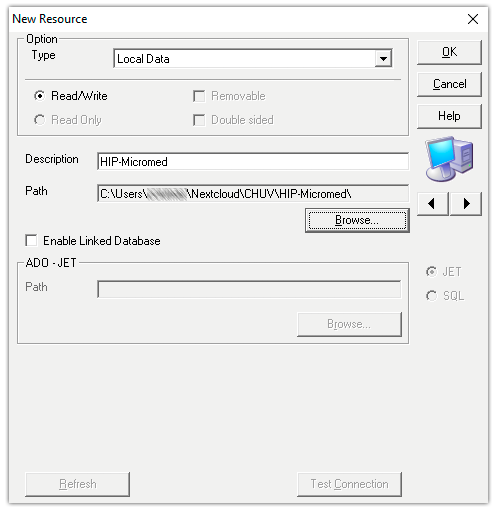
1. Create a new database. In the SystemPLUS EVOLUTION software, go to your System View and create a new Database. the name of the database must be “HIP-Micromed” and the path to the database should be inside the Nextcloud synchronized folder.
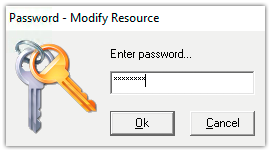
2. Specify the password. The password has to be “micromed” in order to initialize the database.
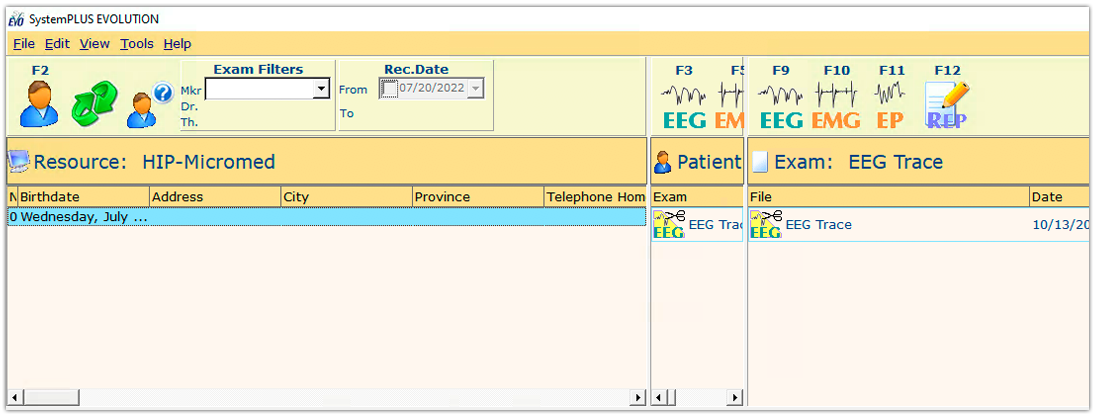
3. Check the new database. Your new database is created and should be visible in the SystemPLUS EVOLUTION software.
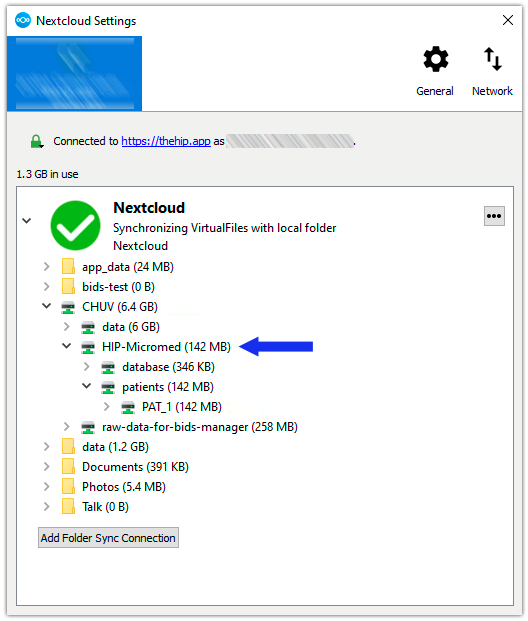
4. Check the synchronization. Check in Nextcloud settings that the newly created “HIP-Micromed” database appears in the synchronized folder.
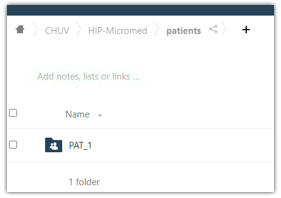
5. Transfer data to the HIP. Any data imported into the “HIP-Micromed” database using the SystemPLUS EVOLUTION software are now accessible in the HIP user’s Private Space.
How to use Collaborative Projects
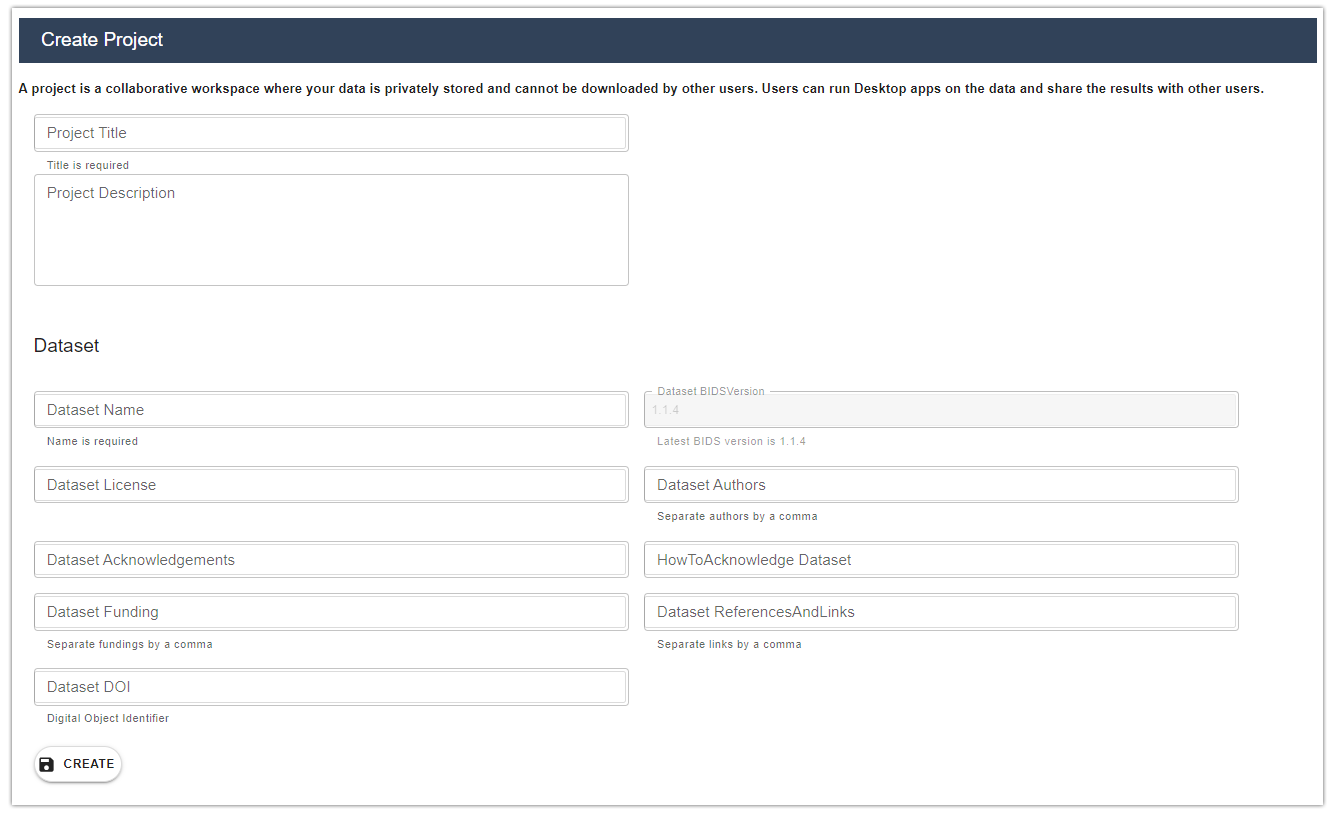
Collaborative Project. Create a new Collaborative Project and share data with HIP users.
About this tutorial
The purpose of this guide is to provide an overview of Collaborative Projects, including their objectives and primary utilization. It comes with a video tutorial demonstrating key operations such as creating a new Collaborative Project, adding members, and transferring and processing data using the integrated tools. It also outlines crucial guidelines for creating and using Collaborative Projects. However, it does not delve into the specific applications of certain services, such as Desktops or BIDS tools, as these are covered in separate guides.
What are Collaborative Projects
A Collaborative Project represents a work group assigned to a research project, contributing through data sharing and/or processing.
The HIP user initiating a Collaborative Project assumes the role of Project Leader, concurrently becoming the Data Controller for that project. The Project Leader holds the authority to add or remove HIP users from any projects under their responsibility.
Members of a project, referred to as Collaborators, are granted access to a set of services enabling them to transfer data and/or documents from their Private Space to the project’s dedicated space. This data transfer can only be performed using HIP’s integrated tools. Once shared, the data can be collectively managed and processed using project-specific BIDS tools and Desktops.
Collaborative projects are the only way to share data between users from different institutions.
Video guide
The following video guide (4’59’’) serves as an introduction to Collaborative Projects: